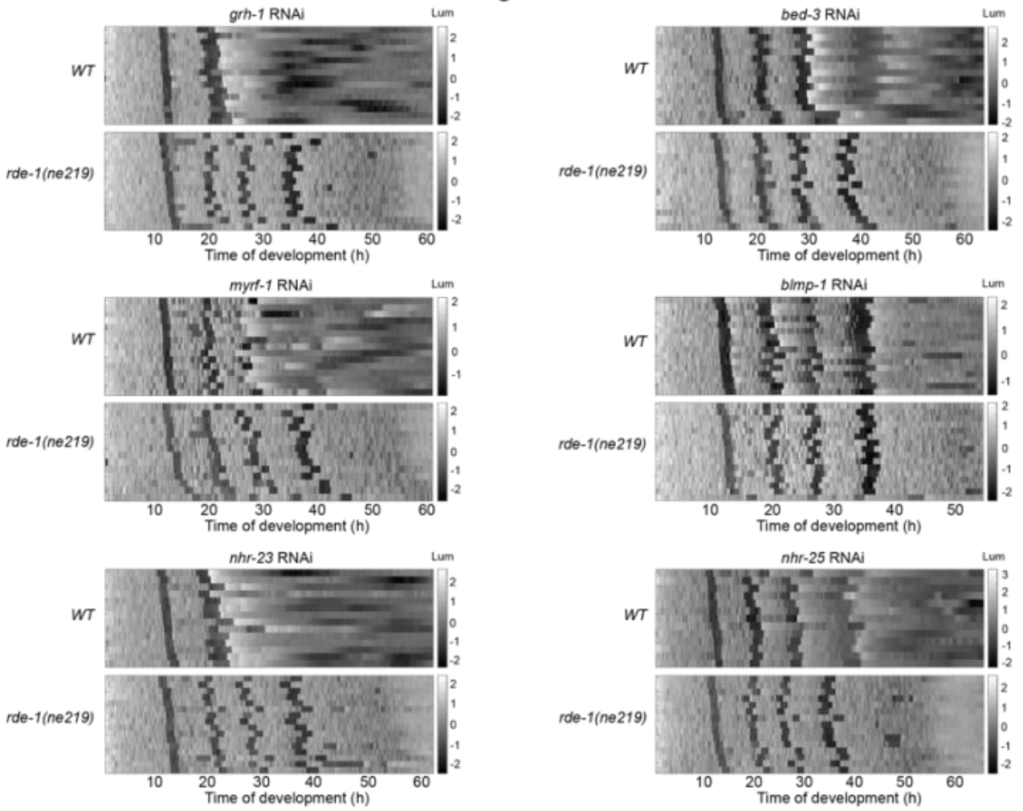
The molting cycle in C. elegans is tightly coupled to the rhythmic accumulation of thousands of transcripts. Here, using ChIP-seq and quantitative reporter assays, we
show that these dynamic gene expression patterns rely on rhythmic transcription. To gain insight into the relevant gene regulatory networks (GRNs), we performed an RNAi-based screen for transcription factors required for molting to identify potential components of a molting clock. We find that depletion of GRH1, BLMP-1, NHR-23, NHR-25, MYRF-1 or BED-3 impairs progression through the molting cycle.
Read our preprint at bioRxiv and a thread with the key findings @LabGrosshans



